This is the story of how the Brodland lab came to have a computational model of neurulation. I hope that it will give you a better understanding of how science is carried out, how scientific knowledge changes with time and how one establishes a research lab. I also hope that it will encourage you in your own work.
G. Wayne Brodland
Why is a "Civil Engineer" studying embryo development?
The answer to this frequently-asked question is quite simple: in 1985, Richard Gordon, a biologist (actually a botanist and radiologist who was studying biology) at the University of Manitoba came to the Faculty of Engineering where I was studying in hopes of finding a collaborator who could help him to build a computational model of neurulation. Neurulation is a critical developmental process in which a sheet of tissue (the neural plate) in an early-stage embryo rolls up to form a tube, the precursor of the spinal cord and brain. At the time, construction of such a model was a very novel idea. I was midway through my Ph.D. under the supervision of Harley Cohen, and having had a bit of a false start, and now having a solid thesis topic well in hand, did not want to change directions. However, I was very interested in the problem and thought that it should take no more than a year or so to modify the large-deformation finite element code I had written as part of my Ph.D. thesis so that it could be used to study this problem. How hard could it be to model a flat sheet of tissue rolling up into a tube!? Richard and I agreed to work together on this problem as soon as I finished my Ph.D., and we did work together for quite a number of years, but not the full 20 (yes, twenty) that it ultimately took to solve the problem. Youth, optimism and inexperience evidently led me to seriously underestimation the complexity of the problem and the work required. I often tell people that, like my Civil Engineering colleagues down the hall, I study structures, it's just that the ones I study are very, very, VERY small.
A 2D model of Neurulation
A bright, young University of Waterloo grad by the name of David Clausi came to my office one day seeking a Master's supervisor, and so began a joint effort to write code to model embryonic tissues at the quasi-cellular level. At least one other model of a cross-section through embryonic tissue existed at the time, but we wanted to build one that as much as possible used only known mechanical effectors: viscous cytoplasm and actin-driven apical constrictions. Ultimately, we were able to write and debug our code and find the conditions under which neurulation would occur. Our model suggested that neurulation was a rather robust process, but that developmental defects could be induced by sufficiently large changes to the forces generated in the neural plate, the thickness of the neural plate or the mechanical resistance offered by the non-neural ectoderm. In this work, we were much encouraged by a series of modest, but crucial, grants from the Spina Bifida and Hydrocephalus Association of Ontario (SBHAO) and later funding from the Easter Seals Research Institute of Ontario.
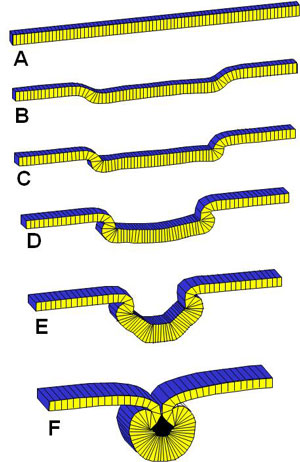
Experimental Data for Validation of the 2D Model

Much of the geometric information we needed to validate the models was not available in the literature, Mike Scott but it was obtained by another bright young UW grad, Mike Scott, who came to do his Master's in our lab. He was an expert at getting people to work together and getting things done, and he worked with a small arm of eager undergraduates who wrote code modules in Pascal (a computer language) under his guidance and mine. He put the code segments together over one Christmas break and the result was a computer program that could actually run a microscope under computer control - quite an accomplishement at the time. The code ran a stepper motor connected to the focus knob of our highly-prized Wild M420 research microscope (at the time, it took a good chunk of our annual budget to purchase it). Our motorized microscope took images at different focal planes, figured out which parts of each image were in best focus and produced a single in-focus collage. The images were a big improvement over what was available at the time
A failed attempt to model in 3D
When we attempted to generalize David's 2D model to 3D, we got stuck. Eventually we realized that embryonic tissues had certain mechanical properties that we did not yet understand. Somehow, they were more fluid in-plane than are standard materials. The problem was especially evident at the cephalic end of the neural plate where the plate rolled inward from each side and from the cephalic end at the same time without wrinkling. When we tried to do this with a sheet of paper, it wrinkled; and when we tried to do it with our computational model, it stalled. Clearly there was something about the mechanics of cells in sheets that we did not understand.
Building a cell-level model
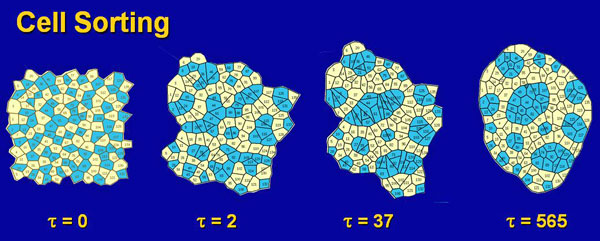
To better understand the mechanics of cell sheets (epithelia), we developed a cell-level model. This demanding work was done by Helen Hong Chen, an ambitious and determined Ph.D. student. Helen Chen One of the big challenges of her work was to figure out how to code the rearrangement that occurred as cells changed neighbours. This was a type of remeshing problem and at the time was quite novel. Once she had figured out how to deal with the extensive "bookkeeping" and finite element challenges involved and got her code running, we needed a way to validate her code. I often tell my students, "assume that computer code is wrong until proven otherwise." We decided to test her code on cell sorting, since this was a biological problem that had been studied extensively for at least the previous 30 years. Basically, if two types of embryonic cells are dissociated and then mixed and reaggregated, the individual cells move spontaneously over time, ultimately producing a compact mass of one type surrounded by the cells of the other type. "Everyone knows how cell sorting works," and so it was a good problem against which to test our code.
A big surprise
When we built 2D aggregates containing 2 types of cells, we found that our model cells invariably sorted backwards compared to what the literature stated. That is to say the wrong type of cells moved to the outside of the mass. This was a big problem, and we knew that a coding error as simple as a minus sign where there should be a plus sign somewhere in our thousands of lines of computer code would be enough to cause it. So we began a careful series of tests to try to find the mistake in our code. Six months of work did not reveal the expected error. It was a very challenging time for us. Eventually, we went to the library to consult a book on interfacial tensions and related phenomena. In one of the books we found there we discovered that when adhesions exist between two immiscible fluids, the surface tensions are LOWERED, not increased. This knowledge told us that our code was in fact correct, and that the traditional interpretation of the many beautiful cell sorting experiments in the literature was not. We had great difficulty publishing our radical work, but eventually ASME agreed to publish one of our papers. With our foot in the door, so to speak, it became increasingly easy to advance this new perspective. Through a series of studies and papers, we eventually came to a general theory of how cells in aggregates rearrange themselves, and our work became more broadly accepted.
3D experimental studies of neurulation
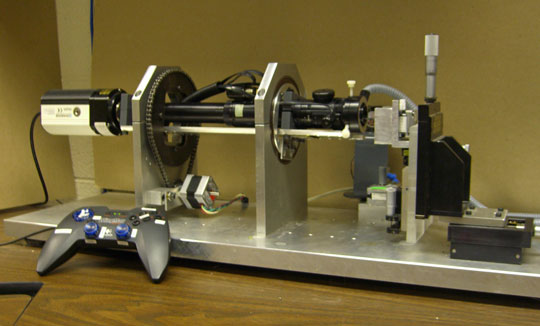
While these cell-level studies were underway, we began detailed experimental studies of neurulation in amphibians (axolotls). Frogatron 3000 A robotic microscope, developed by Jim Veldhuis and other mechanically inclinded students, that we eventually named the "Frogatron 3000", was the key to these studies. It collected time-lapse images from multiple viewing angles over the surface of an embryo and made possible precise 3D reconstructions of the surfaces of live embryos. Tissue motions could be tracked accurately, strain rates calculated and mapped with respect to time and location, and cell fabric mapped.
Mechanical data of embryonic epithelia
Partly to validate our cell-level models and partly so that we would have good mechanical data for our still-planned 3D model of neurulation, Colin Wiebe we developed a device to measure the mechanical properties of embryonic epithelia. After many tries, Colin Wiebe a talented, mechanically-minded mechanical engineer and others in our lab came up with a way to stretch pieces of embryonic tissue as small as 300 microns in size and to measure the applied loads. The stiffness of embryonic tissue samples were found to depend on tissue type, the direction of elongation (they were anisotropic), location within a tissue (they were inhomogeneous) and stage of development (they were time-dependent).
A 3D Model

Finally, it seemed, we were in a position to build a 3D model of neurulation. Xiaouguang Chen, a resolute Ph.D. student took on the task. Xiaoguang Chen His model used geometric information from Frogatron reconstructions and serial sections, and mechanical property information from our tensile tests. Mechanical property assignments were informed by studies that had been done by Keller, Wallingford and others that showed the mechanical effects of certain relevant genes. The model was built using "super elements" each of which represented a small region of the embryo consisting of many cells, and whose properties and cellular fabric were calculated using the constitutive equations we had recently developed. In the end, this model was able to reveal the forces that drive normal neurulation and it showed that changes of as little as 20% in the driving forces, tissue properties or embryo geometry are sufficient to interrupt neural closure. This is the objective to which we began our trek some 20 years earlier.
Many other students have made important contributions to our lab, and their contributions might be the subject of another story...