THE BASIC IDEA
Could information about the forces acting in cells be inferred from their geometries?
Figure 1 – The boundariesof the cells in this image are visible as light-coloured lines and curves [Reference 1]
This intriguing question came to me (G. Wayne Brodland) as I was considering how changes in driving forces in our computer simulations gave rise to changes in cell shape.
It took many tries over five years, but eventually – with the help of a dedicated team of students, technicians and collaborators –we were able to develop a reliable mathematical approachthat could infer the forces in cells from their shapes. We use the term “infer” rather than “calculate”, because the technique does not determine the actual values of the forces, but rather determines the ratios of one edge force to another. It is these ratios that determine how various tug-of-war scenarios play out and whether the movements and reshaping of cells in any particular setting will be normal or abnormal.
The analysis showed that the tensions in any one cell can vary substantially from one location to another along its perimeter (Figure 2), and that the tensions in adjacent cells within a single population (the large cells, for example)could vary substantially from one cell to the next. Both of these findings were unexpected, and had implications for our understanding of the forces present in early stage embryos [1].
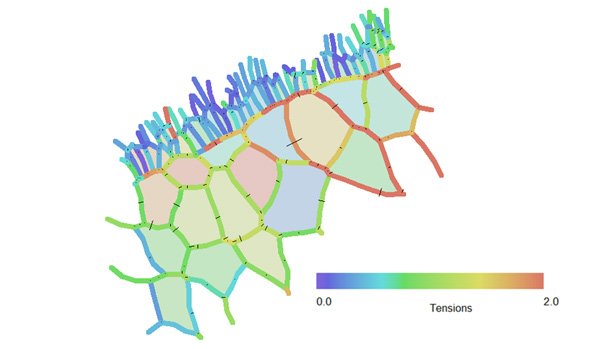
Figure 2 – This image shows the inferred relative tensions of many of the cell edgesshown in Figure 1. Edges shown in blue have relatively low tensions, while those in red have higher ones. [1]
As Video 1 demonstrates, the basic ideas behind this force inference technique are not difficult to understand.
Movie 1 - Inferring Forces in Cells. Download Script
There are now several versions of the technique, including one for cells that are in 2D sheets (as in Figure 1) and another for cells that are in 3D masses [2]. We developed a 2D software suite called theCellular Force Inference Toolkit (CellFIT) [1], and it allows the consistency of the input data to be assessed and the precision of each of the calculated edge tensions can be estimated. A 3D version of the technique has proved useful for predicting the motility of murine breast duct cells [3], a finding that could have relevance to cancer metastasis.
Technical details of the various techniques are available elsewhere [1, 2, 4-6], and are not repeated here.
ABOUT CellFIT
What is CellFIT?
CellFIT is a package of force inference equations and assessment tools that calculates the forces in cell edges based on their geometries. The technique segments the cells in an image and constructs equilibrium equations for each triple junction based on the angles at which the cell edges approach it. Solving the resulting system of tension equations yields a set of relative edge tensions whose scaling must be determined from data external to the image.
The Theory behind CellFIT
The main details of the CellFIT method are presented in References [1] and [6]. Note that CellFITsupercedes an earlier force inference technique that was known as Video Force Microscopy (VFM) [5].
Practical considerations
The CellFITversion available for download here works on planar (2D) data and is ideal for images of cells in monolayers. Images should be taken normal to the surface of the cells. If cells are not in a flat plane (tilted by less than approximately 30 degrees) the images should be optically adjusted to compensate. Additional practical considerations have been detailed elsewhere [6].
CellFIT 3D
We recently developed a 3D version of CellFIT [2]. It was used to determine the tensions acting in exterior and interior interfaces of 8-cell marine embryos. The estimated errors in the exterior tensions were smaller than the 10% uncertainty in companion aspiration experiments. CellFIT-3D has also been used to successfully predict the motility of murine breast duct cells [3].
The computer code that we used to implement CellFIT-3D consisted of several independent software modules, including ones for image processing, trace alignment, angle determination and force analysis. Regrettably, we did not have the resources to amalgamate these codes into a single package, nor did we have the time and data sets needed to validate the software so that it could be released to other users.
We hope that others will pick up the software and the science where we laid them down.
Useful Links
CellFIT co-author M. Shane Hutson.
Seedwater Segmentation Software.
Here you can download a Zipfile that contains a Windows version of Zazu (a 2D version of CellFIT), a User’s Guide and sample data. This software was written with care and was tested on dozens of data sets. However, it is supplied "AS IS" without any warranties or support.
References
[1] Brodland, G.W, Veldhuis, J.H., Kim, S., Perrone, M., Mashburn, D., Hutson, M.S., 2014, "CellFIT: A Cellular Force-Inference Toolkit Using Curvilinear Cell Boundaries," PLoS ONE, Vol. 9, No. 6, e99116. doi: 10.1371/journal.pone.0099116.
[2] Veldhuis JH, Ehsandar A, Maitre J-L, Hiiragi T, Cox S, Brodland GW., 2017, "Inferring cellular forces from image stacks," Phil. Trans. R. Soc. B, 372: 20160261. doi: 10.1098/rstb.2016.0261.
[3] Neil M Neumann, Matthew C Perrone, Jim H Veldhuis, Robert J Huebner, Huiwang Zhan, Peter N Devreotes, G Wayne Brodland, Andrew J Ewald, 2018, “Coordination of Receptor Tyrosine Kinase Signaling and Interfacial Tension Dynamics Drives Radial Intercalation and Tube Elongation”, Developmental Cell, 45, 67-82, e6 doi.org/10.1016/j.devcel.2018.03.011
[4] Cranston, P.G., Veldhuis, J.H., Narasimhan, S. and Brodland, G.W., 2010, "Cinemechanometry (CMM): A Method to Determine the Forces Driving Morphogenetic Movements from Time-lapse Images," Annals of Biomedical Engineering , Vol. 38, pp. 2937-2947. doi: 10.1007/s10439-010-9998-1
[5] Brodland, G.W., Conte, V. Cranston, P.G., Veldhuis, J., Narasimhan, S., Hutson, M.S., Jacinto, A., Ulrich, F., Baum, B., and Miodownik, M., 2010, "Video Force Microscopy Reveals the Mechanics of Ventral Furrow Invagination in Drosophila," Proceedings of the National Academy of Sciences (PNAS), Vol 107, No. 51, pp. 22111-22116. doi: 10.1073/pnas.1006591107 (open access).
[6] Veldhuis, J.H., Mashburn, D., Hutson, M.S., Brodland, G.W., 2015, "Practical aspects of the cellular force inference toolkit (CellFIT)," Methods in cell biology, Vol. 125, pp. 331-351. doi:10.1016/bs.mcb.2014.10.010. preprint pdf.